Software at CIS : lddmm-similitude and lddmm-rigid
Algorithm
Let
![]() and
and
![]() be corresponding template
and target landmarks respectively. Find the rotation,
be corresponding template
and target landmarks respectively. Find the rotation, ![]() ,
translation
,
translation ![]() , and (for lddmm-similitude) scale
, and (for lddmm-similitude) scale ![]() which minimize
which minimize
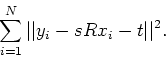
For lddmm-rigid, scale ![]() is fixed at 1.0.
is fixed at 1.0.
Reference
S. Umeyama, "Least-Squares Estimation of Transformation Parameters Between Two Point Patterns".IEEE Transactions on Pattern Analysis and Machine Intelligence, Vol 12, NO 4, April 1991. (pdf)
lddmm-similitude matching is a program to determine the similarity between two landmark data sets based on rotation, translation, and scale. The program will write the rotation, translation and scale to standard out and create a file representing the transformed template.
lddmm-rigid matching is a program to determine the similarity between two landmark data sets based on rotation and translation. The program will write the rotation and translation to standard out and create a file representing the transformed template.
Typical Usage and Parameters:
lddmm-similitude || lddmm-rigid [OPTIONS] x_file y_file tx_file norm_constant [Rts_file]
The following are the command line options.
- x_file: the template landmark file
- y_file: the target landmark file
- tx_file: the output file containing the transformed template. Use /dev/null if this is not required.
- norm_constant: always use 1.0
- Rts_file: omit this option
- Options:
- -r --allowReflection: allow a transform that includes a reflection
- -a --all-info: print data provenance info
- -x --all-info-xml: print data provenance info in an xml format
- -v --version: print program version information
- -h --help: print program usage information
The format for a 2-D landmark file is as follows:
And for a 3-D file:
Output File Format:
The program will format the output file based on the extension of the output
file:
- bz - blitz++ (or runSim) format, described above.
- lmk - BrainWorks format.
- fcsv - Slicer 3D fiducial format.
Here is an example Perl script that computes the similarity for every pair of landmark sets in start_folder and writes the output line by line to a text file for later analysis. This can easily be adapted to do different comparisons (ex. left & right data separately) by calling the function do_sims on a number of different lists:
#!/usr/bin/perl
$program = "lddmm-similitude";
$start_folder = "/cis/project/botteron/hippocampus/Converted_Lmk";
@inlist= `ls -1 $start_folder/*.lmk`;
$outfile = "/cis/project/botteron/hippocampus/left_scale_list.txt";
&do_sims ($infile, \@outlist);
sub do_sims {
my $outfile = shift;
my $list_ref=shift;
my @list = @$list_ref;
print "$outfile\n";
print "List size: $#list\n";
for($i=0; $i<=$#list; $i++) {
chomp $list[$i];
$list[$i] =~ /^.+\/(.+)/;
$shortlist[$i] = $1;
}
open OUT, "> $outfile";
for($i=0; $i<=$#list; $i++) {
for($j=$i+1; $j<=$#list; $j++) {
$sim = `$program $list[$i] $list[$j] /dev/null 1.0 | cut -f3 -d\\ `;
print OUT "$shortlist[$i]:$shortlist[$j]:$sim";
}
print "Finished $shortlist[$i]\n";
}
close OUT;
}
Software developed with support from National Institutes of Health NCRR grant P41 RR15241.
Last Modified: Friday, 15th April, 2011 @ 11:12am






