Research
Researchers at the Center for Imaging Science are developing systems that can interpret images of natural scenes, for example ordinary indoor and outdoor photographs, CT scans and other data obtained with bio-medical imaging devices, and aerial and satellite images acquired by remote sensing. Though great advances have been made in the acquisition of image data, e.g., the development of cameras and other imaging devices, and though the semantic understanding of the shapes and other objects appearing in images is effortless for human beings, the corresponding problem in machine perception, namely automatic interpretation via computer programs, remains a major open challenge in modern science. In fact, there are very few systems whose value derives from the analysis rather than production image data, and this “semantic gap” impedes scientific and technological advances in many areas, including automated medical diagnosis, industrial automation, and effective security and surveillance.
Medical Image Analysis 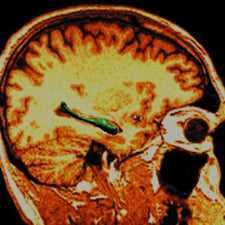
Due to the rapid development of imaging sensor technologies, investigators in the physical and biological sciences are now able to observe living systems and measure both their structural and functional behavior across many scales, from global, aggregate behavior to the microscopic scale of sub-cellular structure. Combining biomedical imaging science with computational modeling, we are now able to infer, noninvasively, the structural and functional properties of complex biological systems and neural circuits, for instance study the cohorts of neuropsychiatric illnesses including schizophrenia, depression, epilepsy, dementia of the Alzheimer type, and Parkinson’s.
Faculty: Barta, Miller, Priebe, Prince, Ratnanather, Sulam, Vidal, Younes
Access the Computational Anatomy Portal
Computer Vision 
Humans annotate scenes much better than computers. With apparently no effort, we readily distinguish among a remarkable variety of objects, actions and interactions in complex, cluttered scenes, e.g., recognizing subtle behaviors among a group of people interacting with each other and with everyday objects. In contrast, automatically interpreting such scenes has proved surprisingly resistant to decades of research. Researchers at CIS are developing algorithms for the automatic interpretation of complex scenes. Examples include detecting faces and deformable objects (e.g. cats) in photographs, recognizing object categories in photographs, segmenting and tracking of moving objects in a video, recognizing dynamic textures (water, smoke, fire) in a video, recognizing human activities in a video, and modeling and recognizing skill in surgical motion and video data.
Faculty: Geman, Hager, Sulam, Tran, Vidal, Younes
Computational Biology 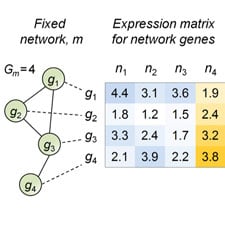
Researchers at CIS are developing new stochastic models and computational methods to analyze regulatory networks, including intracellular signaling and transcription regulation in single cells. One objective is to provide a stochastic representation of the time evolution of molecular copy numbers in cells and to construct novel mathematical and computational tools for their analysis. Other work involves learning graphical models to capture the statistical dependency structure among gene and protein expression values, especially in the “small-sample” regime.
Statistical Learning 
Very high-dimensional data sets are ubiquitous in computational vision, speech and biology. In particular, the past few years have witnessed an explosion in the availability of data from multiple sources and modalities, such as images and videos, genomics data, proteomics data, magnetic resonance images (MRI) and diffusion-weighted images. Unfortunately, the automatic interpretation of such data raises serious challenges in statistical learning. The difficulties are especially pronounced when the objective is to uncover a complex statistical dependency structure within a large number of variables and yet the number of samples available for learning is relatively limited. Researchers at CIS are developing algorithms based on graphical models for reducing the dimensionality of high-dimensional datasets, learning from very few examples, clustering data living in multiple subspaces and manifolds, and discovering relationships in high-dimensional datasets.
Faculty: Geman, Priebe, Sulam, Vidal, Younes

